SEARCH FOR NEW TYROSINE KINASE INHIBITORS AMONG 2-(3-R-1H-1,2,4-TRIAZOL-5-YL)ANILINES AS POTENTIAL ANTITUMOR AGENTS USING MOLECULAR DOCKING
DOI:
https://doi.org/10.15421/jchemtech.v31i2.284813Keywords:
2-(3-R-1,2,4-triazol-5-yl)anilines; tyrosine kinase inhibitors; molecular docking; non-small cell lung cancer; antitumor agents.Abstract
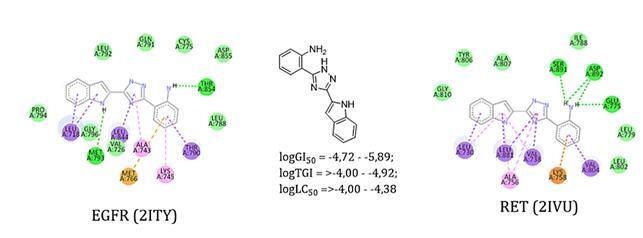
The present work is devoted to the in silico study of 2-(3-R-1H-1,2,4-triazol-5-yl)anilines as potential inhibitors of EGFR (epidermal growth factor receptor) and RET (rearranged during transfection)-, which play a significant role in regulating the physiological cycle of non-small cell lung cancer. The well-known docking software AutoDock Vina was used for the study. The seven studied compounds and two standard drugs (vandetanib and gefitinib) were docked to the crystal structures of EGFR and RET proteins. It was found that among the newly investigated substances, 2-(3-(indolyl-2)-1H-1,2,4-triazol-5-yl)aniline (a5) has the highest affinity towards EGFR and RET with the binding energy of –9.7 and –8.7 kcal/mol, respectively. Visualization of the molecular docking results of this compound using the Discovery Studio software showed that it is characterized by similar to standard ligands location in the active sites of the enzymes, stable hydrogen bonds and π-stacking interactions, which are provided by the presence of indole and aniline fragments in the molecule. Thus, we have identified a new effective ligand that can be used as a "base" molecule for further fragment-oriented design using molecular hybridization methodology (fragment fusion, coupling or extension methods) or structural modification by introducing "pharmacophore" groups into the molecule. Summarizing the above screening results, we can say that 2-(3-R-1H-1,2,4-triazol-5-yl)anilines require further careful consideration as effective tyrosine kinase inhibitors for the search for promising anticancer agents for the treatment of non-small cell lung cancer.
References
Khan, I., Garikapati, K. R., Setti, A., Shaik, A. B., Makani, V. K. K., Shareef, M. A., Rajpurohit, H., Vangara, N., Pal-Bhadra, M., Kamal, A. (2019) Design, synthesis, in silico pharmacokinetics prediction and biological evaluation of 1, 4-dihydroindeno[1,2-c]pyrazole chalcone as EGFR/Akt pathway inhibitors. Eur J Med Chem, 163, 636–648. https://doi.org/10.1016/j.ejmech.2018.12.011.
Tsim, S., O’dowd, C. A., Milroy, R., Davidson, S. (2010). Staging of non-small cell lung cancer (NSCLC): a review. Respiratory medicine, 104(12), 1767–1774. https://doi.org/10.1016/j.rmed.2010.08.005.
Evison, M., AstraZeneca, U. K. (2020). The current treatment landscape in the UK for stage III NSCLC. British Journal of Cancer, 123(Suppl 1), 3–9. https://doi.org/10.1038/s41416-020-01069-z.
Kong, L. L., Ma, R., Yao, M. Y., Yan, X. E., Zhu, S. J., Zhao, P., Yun, C. H. (2017). Structural pharmacological studies on EGFR T790M/C797S. Biochemical and Biophysical Research Communications, 488(2), 266–272. https://doi.org/10.1016/j.bbrc.2017.04.138.
Ibrahim, M. T., Uzairu, A., Shallangwa, G. A., Uba, S. (2021). Computer-aided design of some quinazoline analogues as epidermal growth factor receptor inhibitors. Egyptian Journal of Medical Human Genetics, 22(62), 1–10. https://doi.org/10.1186/s43042-021-00181-w.
Yarden, Y., Sliwkowski, M. X. (2001). Untangling the ErbB signalling network. Nature reviews Molecular cell biology, 2(2), 127–137. https://doi.org/10.1038/35052073.
Schlessinger, J. (2004). Common and distinct elements in cellular signaling via EGF and FGF receptors. Science, 306 (5701), 1506–1507/. https://doi.org/10.1126/science.1105396.
Chan, B.A., Hughes, B.G. (2015) Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Translat Lung Cancer Res. 36, 4(1), 36–54. https://doi.org/10.3978/j.issn.2218-6751.2014.05.01.
Zhang, Q., Wang, Z., Guo, J., Liu, L., Han, X., Li, M, Fang, S., Bi, X., Tang, N., Liu, Y. (2015). Comparison of single-agent chemotherapy and targeted therapy to first-line treatment in patients aged 80 years and older with advanced non-small-cell lung cancer. Onco Targets Ther., 8, 893–898. https://doi.org/10.2147/OTT.S81837.
Wang, X. (2013). Structural studies of GDNF family ligands with their receptors—Insights into ligand recognition and activation of receptor tyrosine kinase RET. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1834(10), 2205–2212. https://doi.org/10.1016/j.bbapap.2012.10.008.
Drilon, A., Hu, Z. I., Lai, G. G., Tan, D. S. (2018). Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nature reviews Clinical oncology, 15(3), 151–167. https://doi.org/10.1038/nrclinonc.2017.175.
Kato, S., Subbiah, V., Marchlik, E., Elkin, S. K., Carter, J. L., Kurzrock, R. (2017). RET Aberrations in Diverse Cancers: Next-Generation Sequencing of 4,871 Patients RET Aberrations in Cancer. Clinical Cancer Research, 23(8), 1988-1997. https://doi.org/10.1158/1078-0432.CCR-16-1679.
Gainor, J. F., Shaw, A. T. (2013). Novel targets in non‐small cell lung cancer: ROS1 and RET fusions. The oncologist, 18(7), 865–875. https://doi.org/10.1634/theoncologist.2013-0095.
Abourehab, M. A., Alqahtani, A. M., Youssif, B. G., Gouda, A. M. (2021). Globally approved EGFR inhibitors: Insights into their syntheses, target kinases, biological activities, receptor interactions, and metabolism. Molecules, 26(21), 6677 https://doi.org/10.3390/molecules26216677.
Bhatia, P., Sharma, V., Alam, O., Manaithiya, A., Alam, P., Kahksha, Alam M. T., Imran, M. (2020). Novel quinazoline-based EGFR kinase inhibitors: A review focussing on SAR and molecular docking studies (2015-2019). European Journal of Medicinal Chemistry, https://doi.org/10.1016/j.ejmech.2020.112640.
Das, D., Hong, J. (2019). Recent advancements of 4-aminoquinazoline derivatives as kinase inhibitors and their applications in medicinal chemistry, European Journal of Medicinal Chemistry, 170, 52–57. https://doi.org/10.1016/j.ejmech.2019.03.004.
Slobbe, P., Windhorst, A. D., Stigter-van Walsum, M., Smit, E. F., Niessen, H. G., Solca, F., Stehle, G., Guus van Dongen, A. M. S., Poot, A. J. (2015). A comparative PET imaging study with the reversible and irreversible EGFR tyrosine kinase inhibitors [11C] erlotinib and [18F] afatinib in lung cancer-bearing mice. EJNMMI Res, 5, 14 https://doi.org/10.1186/s13550-015-0088-0.
Guo, Q., Liu, L., Chen, Z., Fan, Y., Zhou, Y., Yuan, Z., Zhang, W. (2022). Current treatments for non-small cell lung cancer. Frontiers in Oncology, 12. https://doi.org/10.3389/fonc.2022.945102.
Xiaoxia, L., Qian, Y., Pan, W., Changliang, H., Lizi, Y., Funeng X., Zhongqiong, Y., Guizhou, Y., Yuanfeng, Z., Lixia, L., Xu, S., Cheng, L., Wei, Z., Bo, J. (2021). The synthesis review of the approved tyrosine kinase inhibitors for anticancer therapy in 2015–2020 . Bioorganic Chemistry, 113, 105011. https://doi.org/10.1016/j.bioorg.2021.105011.
Bronte, G., Ulivi, P., Verlicchi, A., Cravero, P., Delmonte, A., Crinò, L. (2019). Targeting RET-rearranged non-small-cell lung cancer: future prospects, Lung Cancer: Targets and Therapy, 27–36. https://doi.org/10.2147/LCTT.S192830.
Ferrara, R., Auger, N., Auclin, E., Besse, B. (2018). Clinical and translational implications of RET rearrangements in non–small cell lung cancer. Journal of Thoracic Oncology, 13(1), 27–45. https://doi.org/10.1016/j.jtho.2017.10.021.
Ramesh, P., Veerappapillai, S. (2022). Designing novel compounds for the treatment and management of RET-positive non-small cell lung cancer-fragment based drug design strategy. Molecules, 27(5), 1590. https://doi.org/10.3390/molecules27051590.
Subbiah, V., Yang, D., Velcheti, V., Drilon, A., Meric-Bernstam, F. (2020). State-of-the-art strategies for targeting RET-dependent cancers. J Clin Oncol., 38(11), 1209–21. https://doi.org/10.1200/JCO.
Knowles, P. P., Murray-Rust, J., Kjær, S., Scott, R. P., Hanrahan, S., Santoro, M., Ibáñez, C. F., McDonald, N. Q. (2006). Structure and chemical inhibition of the RET tyrosine kinase domain. Journal of biological chemistry, 281(44), 33577–87. https://doi.org/10.1074/jbc.M605604200.
Yun, C. H., Boggon, T. J., Li, Y., Woo, M. S., Greulich, H., Meyerson, M., Eck, M. J. (2007). Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer cell, 11(3), 217–227. https://doi.org/10.1016/j.ccr.2006.12.017.
Guodong, Z., Shenqian, X., Wuxia, L., Tingting, D., Jingfeng, Z., Minyu, L., Chen, C., Hong, S. (2021). Deciphering the resistance mechanism of RET kinase mutant against vandetanib and nintedanib using molecular dynamics simulations, Journal of Experimental Nanoscience, 16(1), 278–293. https://doi.org/10.1080/17458080.2021.1970141
Reddy, P. S., Lokhande, K. B., Nagar, S., Reddy, V. D., Murthy, P. S., Swamy, K. V. (2018). Molecular modeling, docking, dynamics and simulation of gefitinib and its derivatives with EGFR in non-small cell lung cancer. Current Computer-Aided Drug Design, 14(3), 246–252. https://doi.org/10.2174/1573409914666180228111433
Bommu, U. D., Konidala, K. K., Pamanji, R., Yeguvapalli, S. (2018). Computational screening, ensemble docking and pharmacophore analysis of potential gefitinib analogues against epidermal growth factor receptor. Journal of Receptors and Signal Transduction, 38(1) 48-60. https://doi.org/10.1080/10799893.2018.1426603.
Liang Z, Li Q. X. (2018). π–Cation interactions in molecular recognition: perspectives on pharmaceuticals and pesticides. J Agric Food Chem.; 66(13), 3315–23. https://doi.org/10.1021/acs.jafc.8b00758.
Ramesh P., Karuppasamy R., Veerappapillai S. (2022). A machine learning-based drug rescheduling strategy to identify potential RET inhibitors against non-small cell lung cancer. Medical Oncology, 40 (1), 56. https://doi.org/10.1007/s12032-022-01924-4
Zhao, B., Xiao, Z., Qi, J., Luo, R., Lan, Z., Zhang, Y., Zhu, W. (2019). Design, synthesis and biological evaluation of AZD9291 derivatives as selective and potent EGFRL858R/T790M inhibitors. European journal of medicinal chemistry, 163, 367–380. https://doi.org/10.1016/j.ejmech.2018.11.069
Cross, D. A., Ashton, S. E., Ghiorghiu, S., Eberlein, C., Nebhan, C. A., Spitzler, P. J., Pao, W. (2014). AZD9291, an Irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in Lung Cancer. Cancer discovery, 4(9), 1046–1061. https://doi.org/10.1158/2159-8290.CD-14-0337.
Tanveer, F., Anwar, M. F., Siraj, B., Zarina, S. (2022). Evaluation of anti‐EGFR potential of quinazoline derivatives using molecular docking: An in silico approach. Biotechnology and Applied Biochemistry, 69(3), 1226–1237. https://doi.org/10.1002/bab.2199
Uma, D. B., Kranthi, K. K., Rishika, P., Suneetha, Y. (2018). Computational screening, ensemble docking and pharmacophore analysis of potential gefitinib analogues against epidermal growth factor receptor, Journal of Receptors and Signal Transduction, 38(1), 48–60. https://doi.org/10.1080/10799893.2018.1426603
Bilyi, A. K., Kovalenko, S. I., Prykhodko, O. B., Emets, T. I. (2013). [Investigation of the rist-stimulating activity of 2-heteryl[1,2,4]tryazolo[1,5-s]quinazolines and products of their nucleophilic degradation]. Zaporizkyi medychnyi zhurnal, 2(77), 83–86. https://doi.org/10.14739/2310-1210.2013.2.15606
Karaman, M. W., Herrgard, S., Treiber, D. K., Gallant, P., Atteridge, C. E, Campbell, B, T, Chan, K, W., Ciceri, P., Davis, M. I., Edeen, P. T. (2008). A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol, 26(1), 127–132. https://doi.org/10.1038/nbt1358
El Khatabi, K., El-mernissi, R., Moukhliss, Y., Hajji, H., Rehman, H. M., Yadav, R., Bouachrine, M. (2022). Rational design of novel potential EGFR inhibitors by 3D-QSAR, molecular docking, molecular dynamics simulation, and pharmacokinetics studies. Chemical Data Collections, 39, 100851. https://doi.org/10.1016/j.cdc.2022.100851
Bilyi, A. K., Kovalenko, S. I., Katsev, A. M., Kholodniak, S. V., Fedotova, O.S. (2012). [Investigation of cytotoxicity and antitumor activity of quinazoline derivatives and its condensed analogues]. Zaporozhskyi medytsynskyi zhurnal, 4(73), 60–65. (In Ukrainian).
Protein Data Bank. https://www.rcsb.org/
Trott, O., Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem., 31(2), 455–61. https://doi.org/10.1002/jcc.21334.
Discovery Studio Visualizer v21.1.20298. Accelrys Software Inc., https://www.3dsbiovia.com
Downloads
Published
Issue
Section
License
Copyright (c) 2023 Oles Honchar Dnipro National University

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Authors reserve the right of attribution for the submitted manuscript, while transferring to the Journal the right to publish the article under the Creative Commons Attribution License. This license allows free distribution of the published work under the condition of proper attribution of the original authors and the initial publication source (i.e. the Journal)
- Authors have the right to enter into separate agreements for additional non-exclusive distribution of the work in the form it was published in the Journal (such as publishing the article on the institutional website or as a part of a monograph), provided the original publication in this Journal is properly referenced
- The Journal allows and encourages online publication of the manuscripts (such as on personal web pages), even when such a manuscript is still under editorial consideration, since it allows for a productive scientific discussion and better citation dynamics (see The Effect of Open Access).